|
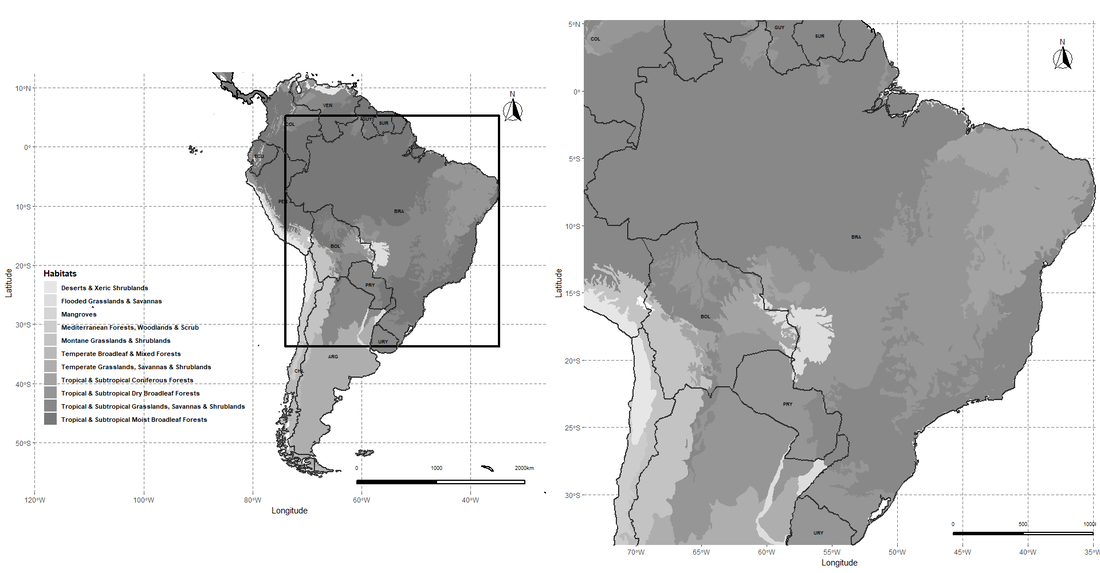
South American ecoregions with a zoom out to Brazil by Russell J. Gray Ecoregions maps are simple in R, here is some reproducible code to make them yourself. You can download the newest version (2017) WWF ecoregion data here code: # make sure to set the working directory to your folder that you have unzipped the the ecoregions shapefile setwd("") library(ggplot2) library(rgdal) library(ggspatial) library(raster) library(cowplot) library(egg) library(rnaturalearth) library(sf) library(hddtools) #------------------------------------------------------------------------ # get a world object with all countries world <- ne_countries(scale = "medium", returnclass = "sf") world.SA <- subset(world, continent=="South America") # get central points and coordinates to label the country names # don't worry about the warning message it just refers to possible # improper location of country name text (but insignificantly so) world_points<- st_centroid(world) world_points <- cbind(world, st_coordinates(st_centroid(world$geometry))) SA <- subset(world_points, continent == "South America") plot(SA) Brazil <- subset(SA, iso_a3 == "BRA") plot(Brazil) #------------------------------------------------------------------------ list.files(pattern=".shp") eco.regions <- readOGR(getwd(), "tm_ecoregions_2017") # get ecoregions for Sout America eco.regions.SA <- subset(eco.regions, REALM=="Neotropic") eco.df <- fortify(eco.regions.SA, region ="BIOME_NAME") #-------------------------------------------------------------- #--------------------------------------------------------------- # create a bounding box around Brazil bbSP <- bboxSpatialPolygon(boundingbox = extent(Brazil)) # fortify the shape file eco.simp <- crop(eco.regions, extent(bbSP)) eco.simp <- fortify(eco.simp, region ="BIOME_NAME") table(eco.df$id) #----------------------------------------------------------------------------------- # Make the map p1 <- ggplot() + geom_spatial_polygon(data = eco.df, aes(x = long, y = lat, group = group, fill=id), alpha = 1, linetype = 1) + scale_fill_manual(values = grey.colors(13, rev = TRUE))+ geom_sf(data = world, fill= NA, lwd=1, color="black") + geom_spatial_polygon(data = bbSP, aes(x = long, y = lat, group = group), size = 1.5, alpha = 1, linetype = 1, color = "black", fill = NA) + geom_sf(data = world.SA, fill= NA, size = 0.5) + geom_text(data= SA,aes(x=X, y=Y, label=wb_a3), size = 2, color = "black", fontface = "bold", check_overlap = FALSE) + annotation_north_arrow(location = "tr", which_north = "true", pad_x = unit(0.3, "in"), pad_y = unit(0.35, "in"), style = north_arrow_fancy_orienteering) + ggsn::scalebar(x.min=-30, x.max=-50, y.min=-57, y.max=55, transform = TRUE, dist = 1000, dist_unit = "km", location = "bottomright", box.fill = c("black", "white"), st.color = "black", height = 0.005, st.bottom = FALSE, st.size = 2.5) + coord_sf(xlim = c(-120, -26.24139), ylim = c(-58.49861, 12.59028), expand = FALSE) + xlab("Longitude") + ylab("Latitude") + ggtitle("") + theme(panel.grid.major = element_line(color = gray(.5), linetype = "dashed", size = 0.5), panel.background = element_rect(fill = "white"), legend.title = element_text(face=2), legend.background = element_blank(), legend.key = element_blank(), legend.position = c(.01, .15), legend.justification = c("left", "bottom"), legend.box.just = "left", legend.text = element_text(size=8,face=2))+ labs(color="Species",fill = "Habitats") p1 p2 <- ggplot() + geom_spatial_polygon(data = eco.simp, aes(x = long, y = lat, group = group, fill=id), alpha = 1, linetype = 1) + scale_fill_manual(values = grey.colors(13, rev = TRUE))+ geom_sf(data = world, fill= NA, lwd=1, color="black") + geom_sf(data = world.SA, fill= NA, size = 0.5) + geom_text(data= SA,aes(x=X, y=Y, label=wb_a3), size = 2, color = "black", fontface = "bold", check_overlap = FALSE) + annotation_north_arrow(location = "tr", which_north = "true", pad_x = unit(0.3, "in"), pad_y = unit(0.35, "in"), style = north_arrow_fancy_orienteering) + ggsn::scalebar(x.min=-35, x.max=-50, y.min=-33, y.max=1, transform = TRUE, dist = 500, dist_unit = "km", location = "bottomright", box.fill = c("black", "white"), st.color = "black", height = 0.005, st.bottom = FALSE, st.size = 2.5) + coord_sf(xlim = c(-74.00205, -34.80547), ylim=c(-33.74219, 5.257959), expand = FALSE) + xlab("Longitude") + ylab("Latitude") + ggtitle("") + theme(panel.grid.major = element_line(color = gray(.5), linetype = "dashed", size = 0.5), panel.background = element_rect(fill = "white"), legend.title = element_text(face=2), legend.background = element_blank(), legend.key = element_blank(), legend.position = "none", legend.justification = c("left", "bottom"), legend.box.just = "left", legend.text = element_text(size=8,face=2))+ labs(color="Species",fill = "Habitats") p2 plot_grid(p1,p2,ncol = 2,nrow = 1)
1 Comment
Xin'e Li
11/28/2023 11:54:32 pm
Very happy to read your blog about plotting ecoregion maps in R (https://www.rjgrayecology.com/blog/ecoregions-maps-in-r#/). Your work helps me a lot! However, I have a question. In your blog, you just plot the Africa, but I need to plot the whole world map filled with the 14 biomes. My main problem is the plotting speed, which is very very slow. Do you have any good ideas to speed up this? I think the dataset is too big, but how to simplify it? I will be very grateful if you can give some advice! thanks!
Reply
Leave a Reply. |
AuthorRussell J. Gray Archives
April 2021
Categories |

 RSS Feed
RSS Feed
